|
| |
Michael Norton and the Norton Group Department of Chemistry, Marshall University |
| |
|
|
|
|
|
|
| |
|
|
|
|
|
|
| |
|
|
| |
ABSTRACT: This presentation serves 3 functions: 1. To provide the reader with an overview of “Directed Sequential Assembly via DNA Based Nanostructures”, one of many possible approaches to nanoelectronic systems, 2. To provide a report of our progress in implementing this approach and 3. To highlight current and potential barriers to implementation of this sequential assembly process. |
|
| |
|
|
|
|
|
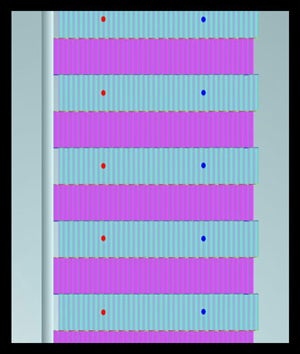
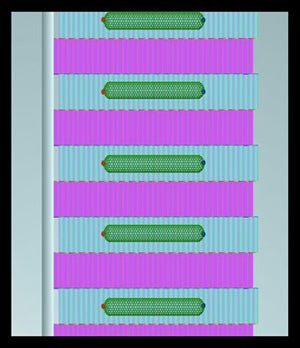
INTRODUCTION: One of the goals of nanotechnology research is the creation of networks of electrically active species which function as integrated systems. The approach to assembly at the nanoscale which the Norton group is implementing is termed Directed Sequential Self-Assembly via DNA Nanostructures. Representations of structures targeted by the group are shown in the animation (see below). The building blocks (color coded violet and cyan) in the animation measure 4 nm by 16 nm. The attachment sites for surface self-assembly (color coded red and blue) are spaced 40 nm (red-blue dot separation) and 32 nm (red dot-red dot or line to line separation). Sequential assembly requires surface immobilization of a growth directing species. The immobilization strategy selected for this application employs the strong interaction between thiols and gold, and is described below. The semibiosynthetic nucleator, which is a one micron long strand of designer DNA is also defined below. |
|
|
|
| |
|
|
|
|
|
|
|
Pictorial representation of a two dimensional asymetcric DNA nanoplatform |
|
|
|
Representation of part of a targeted structure, a two dimensional Nanotube array. |
|
| |
|
 |
|
|
|
|
|
| |
| |
|
|
|
Below are listed 4 set of objectives which the group sought to achieve and the subsequent results. In addition, the challenges which objective presented are indicated.
|
|
| |
| |
|
|
|
|
|
|
|
|
|
|
|
|
Objectives: Prepare a silicon substrate with between 100 and 10,000 pairs of chemically/electrically addressable sites. This will enable parallel simultaneous assembly of multiple copies of the nanostructues, enabling statistical evaluation of the error rates in the slow, sequential assembly process. |
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
| |
|
|
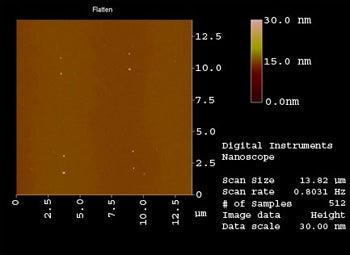
Results: 10 by 10 arrays of 100 nm gold dot pairs has been fabricated by e-beam lithography.
Challenges: Prepare robust 5 nm spots, create electrical interconnects, maintain surface flatness, eliminate processing derived contamination, prevent Si surface fouling during assembly.
|
|
|
|
 |
|
|
| |
|
|
|
|
|
|
|
|
|
| |
| |
|
|
|
|
|
|
|
|
|
|
|
|
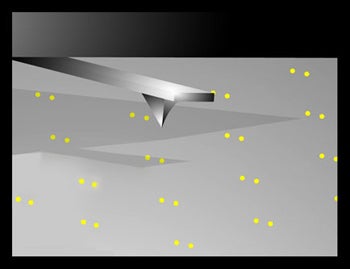
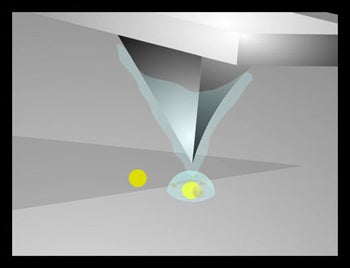
Objectives: Use nanoscale solution deposition to write one side of the director strand to one of the two metallic spots in a dot pair. The resolution is limited not by the diameter of the written spot, but by the dimensions of the metallic dot, enabling sub 20 nanometer scale resolution while writing 50 – 200 nm spots . |
|
|
|
 |
|
|
| |
|
|
|
|
|
|
|
|
|
| |
|
|
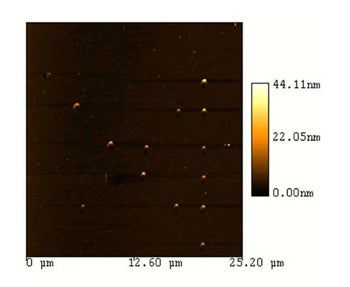
Results: We (and others (2)) have written DNA containing spots with diameters on the order of 250 nm, well below the micron separation between dots.
Challenges: Develop adhesion chemistries or electrochemical methods which generate selective attachment of the director strand to one of each pair of attachment sites |
|
|
|
 |
|
|
| |
|
|
|
|
|
|
|
|
|
| |
| |
|
|
|
|
|
|
|
|
|
| |
|
|
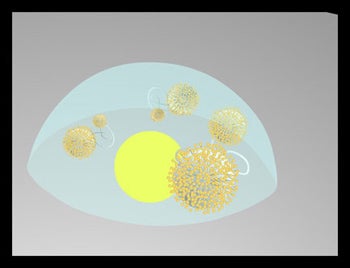
Objectives: Design and synthesize generation 4 thiolated dendrimers monofunctionalized with one 20 mer DNA oligomer which can act as a primer in the Polymerase Chain Reaction (PCR). If the dendrimer diameter closely matches the diameter of the metallic dot, steric factors/competitive binding may limit occupation of the spot to one dendrimer
Results: A “trebler” chemistry is available which allows the production of short DNA molecules terminated with dendrimer like structures with a minimum of 3 thiols. This chemistry is compatible with commercial DNA synthesizers. These reactions, however, are particularly slow and produces low yields (3).
Challenges: Defining an efficient chemical route to large thiolated dendrimers compatible with PCR conditions and identifying suitable methods for characterization of these molecules. |
|
|
|
 |
|
|
| |
|
|
|
| |
| |
|
|
|
|
|
|
|
|
|
| |
|
|
Objectives: Design a repetitive Director (nucleation) Strand, synthesize using rolling circle synthesis (4), clone it into a plasmid, selectively amplify it using PCR, prepare single stranded DNA, attach one or both ends, align using flow, sequentially add building blocks to structure one step at a time.
Results: |
|
|
|
 |
|
|
CTTGACCTGATTCGCCAGCTATTTAGGTGACACTATAGAATACTCAAGCTTGCATGCCTGCAGGTCGACT
CTAGATCGGACAGCAGCCTGACGCTGGTTGCATCGGACGATACTACATGCCAGTTGGACTAACGGCTCT
ACCGTGCATCATGGACTAACCAGTGACCGCATCGGACAGCAGCCTGACGCTGGTTGCATCGGACGATAA
TACATGCCAGTTGGACTAACGGCGCTACCGTGCATCATGGACTAACCAGTGACCGCATCGGACAGCAGC
CTGACGCTGGTTGCATCGGACGATACTACATGCCAGTTGGACTAACGGCGCTACCGTGCATCATGGACT
AACCAGTGACCGCATCGGACAGCAGCCTGACGCTGGTTGCATCGGACGATACTACATGCCAGTTGGACT
AACGGCGCTACCGTGCATCATGGACTAACCAGTGACCGCATCGGACAGCAGCCTGACGCTGGTTGCATC
AGACGATACTACATGCCAGTTGGACTAACGGCGCTACCGTGCATCATGGACTAACCAGTGACCGCATCG
GACAGCAGCCTGACGCTGGTTGCATCGGACGATACTACATGCCAGTTGGACTAACGGGCGCTACCGTGC
ATCATGGACTAACCAGTGACCGCAGAATTCGCCCTATAGTGAGTCGTATTACAATTCACTGGCCGTCGTT
TTACAACGTCGTGACTGGGAAAACCCTGGCGTTAC |
|
Sequencing result. Each copy is depicted in a different color; the red color shows the EcoRI and XbaI restriction sites introduced into the DNA fragment.
Challenges: Cloning multiple tandem repeats and determining the sequence represents a significant challenge. A second challenge involves design of robust building blocks and experimentally identifying failure mechanisms during the sequential assembly process. |
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
| |
| |
|
|
|
|
|
|
|
|
|
| |
|
|
Norton Group and Collaborators
Postdoctoral Fellow: Ashish Vaidya
Graduate Students: Aoune Barhoumi, Fan Zhang, Ava Dykes, Richard Holt, Thabo Gcwabaza
Undergraduate Students: Mikala Shremshock, Fung Chan, Erica Roberts, Justin Swick, Derek Gregg
Technical Support: David Neff, Chris Tucker, Ian Towler, Julie Merrick
Collaborators: Carolyn Matzke (CINT), Joel Wendt (CINT)
|
|
|
|
Literature Sited
1. E. Winfree, E.; Liu, F.; Wenzler, L.A.and Seeman, N.C.: Design and Self-Assembly of Two-Dimensional DNA Crystals, Nature 394, 539-544 (1998).
2. Demers, L.M.; Ginger, D.S.; Park, S.J.; Li, Z; Chung, S.W.; Mirkin, C.A.: Direct Patterning of Modified Oligonucleotides on Metals and Insulators by Dip-Pen Nanolithography Science, 296, 1836-1838 (2002).
3. Li, Z.; Jin, R.; Mirkin, C.A. and Letsinger,R.L.: Multiple thiol-anchor capped DNA–gold nanoparticle conjugates,
Nucleic Acids Research, Vol. 30, No. 7 1558-1562 (2002).
4. Liu,D.; Daubendiek,S.L.; Zillman,M.A..; Ryan, K. and Kool, E.T.: Rolling circle DNA: small circular Oligonucleotides as efficient templates for DNA polymerases. J. Am. Chem. Soc, 118, 1587-1594 (1996).
|
|
|
| |
|
|
|
|
|
|
|
|
|
| |
|
|
. |
|
|
|
|
Click to watch an animation which provides a general overview of the project. You may have to download a free Macromedia Flash Player at www.macromedia.com/downloads. If you have the Flash Player, you may still need to select it manually when prompted to select an application. |
|
|
| |
Back to Example Projects
|